
# Sweep through and compute each new cell row-wise. Result = įor i in range(-1, -len(second_seq) - 1, -1): # Initialize the resulting matrix with the initial row. # Internally requies that the first sequence is longer.įirst_seq, second_seq = second_seq, first_seq
Second_seq : One of the sequences to be compared forĪ 2D list corresponding to the resulting matrix of the Needlem. Generates a matrix corresponding to the scores to the Needleman-Wu-įirst_seq : One of the sequences to be compared for similarity. Oring scheme) what the score for the particular characters can be.ĭef generate(self, first_seq, second_seq): Helper function, given two characters determines (based on the sc. The value for the right bottom corner of a particular block.ĭef _calc_weight(self, first_char, second_char):

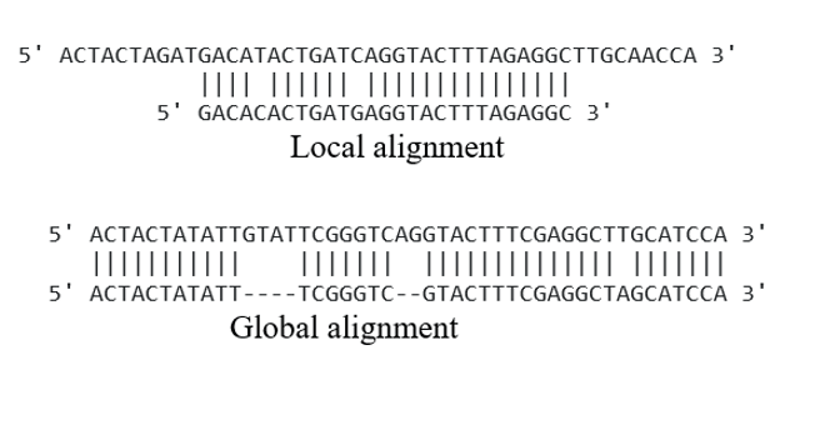
J : The bottom most part of the block being computed. I : The right most part of the block being computed. Result : The current matrix that is being computed. Given a block (corresponding to a 2 x 2 matrix), calculate the value o. needlemanwunsch.py import randomĬlass used for generating Needleman-Wunsch matrices. Currently, the program when ran will generate two random sequences of DNA and then print out the resulting Needleman-Wunsch matrix. I have created a Python program, that given two strings, will create the resulting matrix for the Needleman-Wunsch algorithm. Usually, a grid is generated and then you follow a path down the grid (based off the largest value) to compute the optimal alignment between two sequences. The Needleman-Wunsch algorithm is a way to align sequences in a way that optimizes "similarity".


 0 kommentar(er)
0 kommentar(er)
